Introduction
Lindel is a Logistic regression model for accurate indel prediction induced by Cas9 cleavage. It takes 65 bp sequence (clevage site at 30) as an input and predicts the frequencies for all possible deletions <30 bp, all 1-2 bp insertions, and insertions larger than 2 bp as a group.
In this manual, we are going to introduce how Lindel works and how to use it
Using Lindel
Webtool
We developed a webtool to show the distribution of most frequent indels. This is just a visualization tool, for more functions and detailed information, please install the package locally to generate full list of indels and frameshift ratio predicted by our tool.
Integrated in CRISPOR
Lindel has been integrated into guide design tool CRISPOR.
Installing local Lindel from Github
Lindel has been wrapped as a python package. It includes the functions to parse input sequence and models to predict the frequencies of all indels.
Requirements
Compatible with both Python2.7 and Python3.5 or higher.
Requires biopython,numpy and scipy
In terminal, type:
git clone https://github.com/shendurelab/Lindel.git
cd Lindel
python setup.py install We provide a simple test python code Lindel_prediction.py for you to test your target sequence. It uses 60-65 bp sequence (30 bp upstream and 30-35 bp downstream of the cleavage site) and a filename as input, and outputs the file listing all possible indels and the frameshift ratio.
python Lindel_prediction.py TAACGTTATCAACGCCTATATTAAAGCGACCGTCGGTTGAACTGCGTGGATCAATGCGTC test_seqThe output file should look like the table below, showing the indels, frequencies and locations. The location is relative distance to the cleavage site, with negative values for upstream and non-negative values for downstream.
| ...AACGCCTATATTAAAGCGAC | CGTCGGTTGAACTGCGTGGA... | Frequency | Indel size | Indel start site |
|---|---|---|---|
| ...AACGCCTATATTAAAGCG-- | --TCGGTTGAACTGCGTGGA... | 30.90 | D4 | -2 |
| ...AACGCCTATATTAAAGCGAC | -GTCGGTTGAACTGCGTGGA... | 16.37 | D1 | 0 |
| ...AACGCCTATATTAAAGCGAC X CGTCGGTTGAACTGCGTGGA... | 8.77 | I3+X | |
| ...AACGCCTATATTAAAGCGAC C CGTCGGTTGAACTGCGTGGA... | 6.07 | I1+C | |
| ...AACGCCTATATTAAAGCG-- | -----GTTGAACTGCGTGGA... | 3.96 | D7 | -2 |
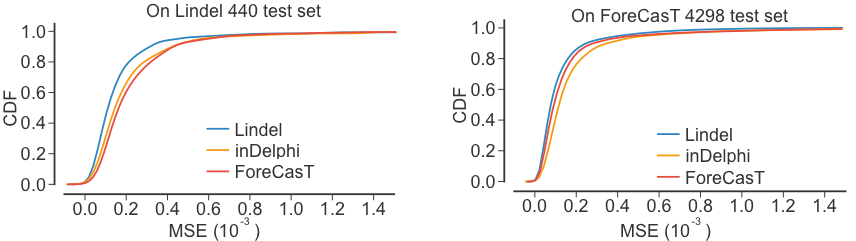
Lindel Performance
Two other tools (ForeCasT and inDelphi) are also available to predict the relative frequencies of indels. Lindel compares favorably to the other tools.